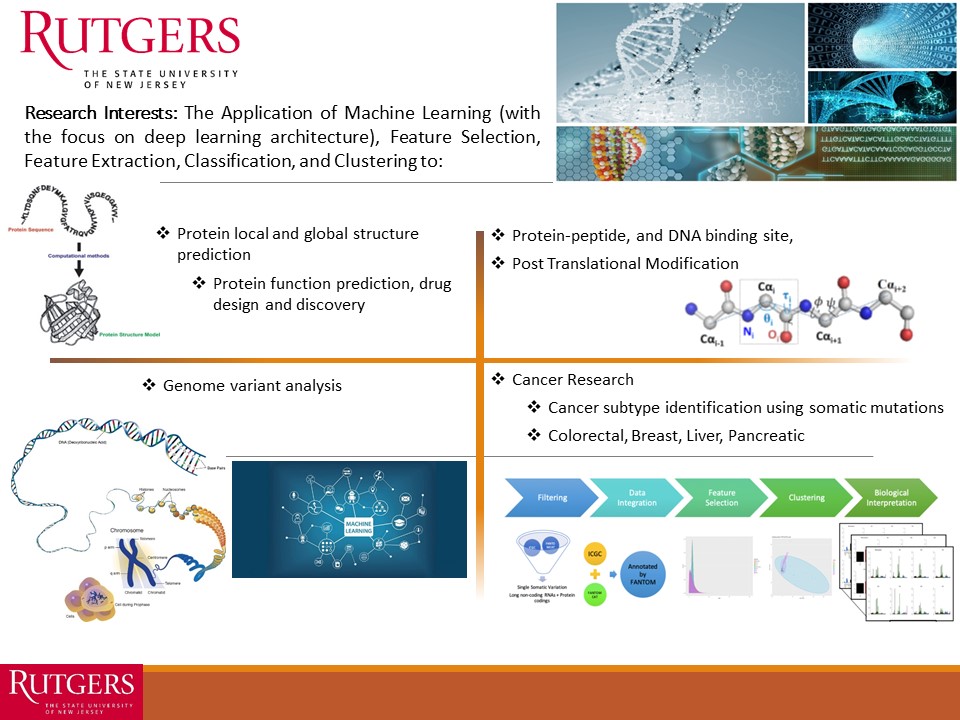
Our research focuses at MLBC are presented in the following quad chart:
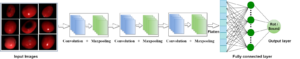
Our research focuses at MLBC are on machine learning, artificial intelligence, and bioinformatics & computational biology in general. We work on the challenges associated with the design and development of robust, general, and accurate systems for several important problems in bioinformatics and computational biology such as, protein local and global structure prediction, genome variants analysis, cancer subtype classification, and studying the impact of somatic mutations in cancer research, in specific. Our focus is on using machine learning techniques with an emphasis on deep learning architecture, support vector machines, and ensemble classifiers (homogeneous and heterogeneous).
USDA Collaboration – Cranberry Harvest Automization
At the MLBC lab, we are proud to collaborate with the United States Department of Agriculture. Together, we are exploring how Machine Learning can innovate agricultural practices in New Jersey. Currently, we are focused on developing methods to develop biological insights and improving existing methods related to cranberry cultivation. More information about this collaboration, along with available code and datasets, can be found at USDA Collaboration.